Overview
NanoDSF stands for the nano-format of Differential Scanning Fluorimetry (DSF). It is a fast, robust, high-quality, and – most importantly – label-free and in-solution method for the analysis of protein stability, thermal protein unfolding and melting temperature analysis. Consequently, nanoDSF is a great tool for buffer and formulation screening as well as screening of small molecule compound libraries for influence on protein stability and shifts of thermal melting temperature. Additionally, nanoDSF allows for analyzing the colloidal stability of protein solutions (aggregation).
In general, the intrinsic tryptophan fluorescence of proteins is strongly dependent on their 3D-structure and hence the local surroundings of the tryptophan residues. Using chemical denaturants or a thermal gradient, proteins can be unfolded, which leads to changes in their intrinsic tryptophan fluorescence. This translates into fluorescence emission peak shifts and intensity changes. NanoDSF monitors these fluorescence changes with high time-resolution and can reveal even multiple unfolding transitions. NanoDSF is therefore highly successful in antibody engineering, membrane protein characterization, protein quality control, buffer screening, protein unfolding analysis, and small molecule compound binding screening.
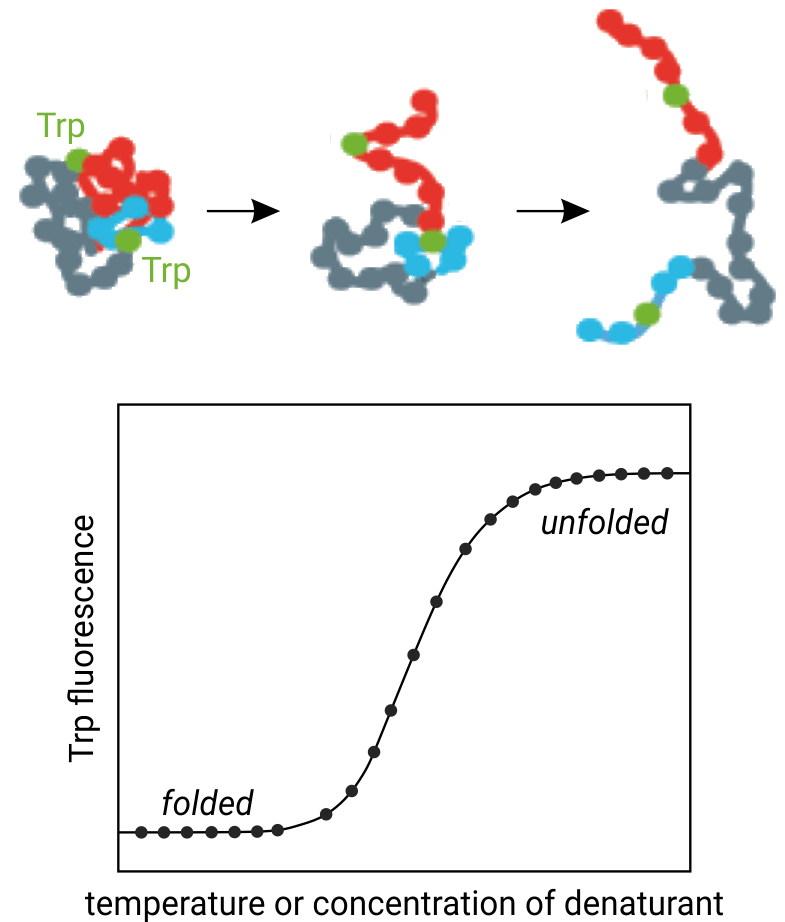
The figure above illustrates the principle behind thermal protein unfolding: Increasing temperature causes unfolding of the three-dimensional protein structure and thus tryptophan residues to become solvent exposed. NanoDSF monitors the concurrent changes in tryptophan fluorenscence at 330 and 350 nm wavelength.
In order to detect protein aggregation, the nanoDSF device features also back-reflection optics. Normally, visible light passes through the capillaries containing the protein sample of interest without any interference, is reflected by a mirror on the capillary tray, and finally quantified by the detector. If the protein sample contains aggregated particles, the incident light is scattered by these particles. The loss of reflection intensity is a precise measure for protein aggregation.
Technology
NanoDSF is a differential scanning fluorimetry method able to analyze the conformational stability and colloidal stability (aggregation behavior) of proteins under different thermal and chemical conditions. The conformational stability of a protein is described by its unfolding transition midpoint Tm (°C), which is the point where half of the protein is unfolded. The truly label-free nanoDSF technique monitors the intrinsic tryptophan fluorescence of proteins, which is highly sensitive for the close surroundings of the tryptophan residues and which changes upon thermal unfolding.
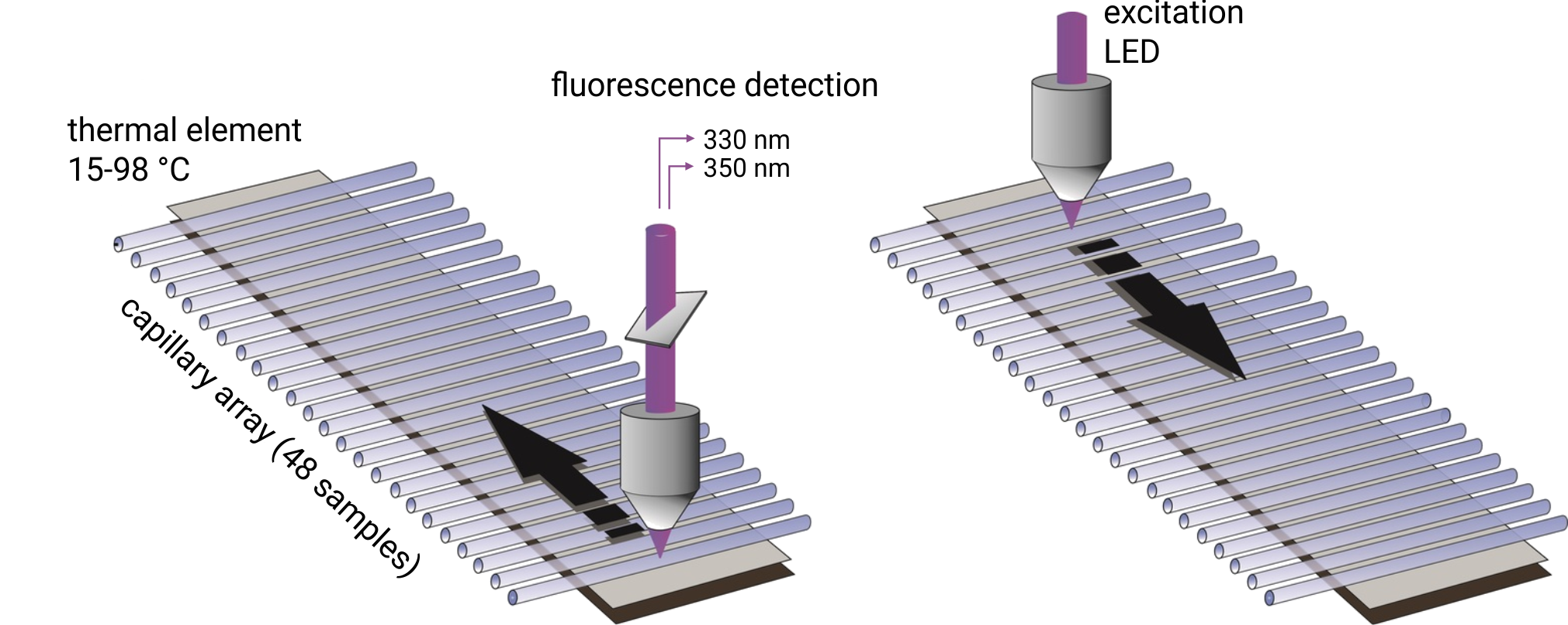
Up to 48 capillaries are filled with 10 µl of protein sample and simultaneously scanned at 330/350 nm wavelengths. Melting temperatures are recorded by monitoring changes in the intrinsic tryptophan fluorescence and aggregation onset temperatures are detected via back-reflection light scattering. The samples can be heated to any temperature in the range from 15°C to 95°C. Importantly, samples can be studied without the use of a dye and with free choice of buffer and detergent. Melting temperatures of proteins with a concentration between 5 µg/ml and 250 mg/ml can be analyzed. In order to obtain high quality aggregation onset temperatures, protein solutions with concentrations above 1 mg/ml are required.
Typical applications
Due to the robust detection method and quick analysis pace, nanoDSF is very versatile and can be used in a number of contexts:
• Screening of buffers, formulation, and buffer additives
• Screening of detergents
• Analysis of thermal and chemical protein unfolding
• Long-term protein and antibody storage optimization
• Forced-degradation stability testing
• Comparison of biosimilar proteins and antibodies with respect to stability and aggregation
• Batch-to-batch comparison assays
• Deep feature analysis (influence of mutations, modifications, conjugations on protein stability and aggregation)
Advantages
• nanoDSF does not require fluorescent dyes like Sypro Orange like in standard DSF (thermofluor)
• Low sample consumption → Only 10 µL of sample required
• Free choice of assay buffers → additives/detergents
• Very short analysis time → enables high throughput
• Optimal data quality and resolution → Dual 350/330 nm UV-detection
• Wide temperature range → Analysis possible from 15°C to 95°C
• No labeling required → Close-to-native analysis possible
• Wide concentration range → 5 µg/ml to 200 mg/ml
• Wide molecule size range → From 1 kDa to 1 MDa